Water bodies detection
Water bodies detection
This application takes as input Copernicus Sentinel-2 or USSG Landsat-9 data and detects water bodies by applying the Otsu thresholding technique on the Normalized Difference Water Index (NDWI).
The NDWI is calculated with:
Typically, NDWI values of water bodies are larger than 0.2 and built-up features have positive values between 0 and 0.2.
Vegetation has much smaller NDWI values, which results in distinguishing vegetation from water bodies easier.
The NDWI values correspond to the following ranges:
| Range | Description |
|---|---|
| 0,2 - 1 | Water surface |
| 0.0 - 0,2 | Flooding, humidity |
| -0,3 - 0.0 | Moderate drought, non-aqueous surfaces |
| -1 - -0.3 | Drought, non-aqueous surfaces |
To ease the determination of the water surface/non water surface, the Ostu thresholding technique is used.
In the simplest form, the Otsu algorithm returns a single intensity threshold that separate pixels into two classes, foreground and background. This threshold is determined by minimizing intra-class intensity variance, or equivalently, by maximizing inter-class variance:

The application can be used in two modes:
- take a list of Sentinel-2 STAC items references, applies the crop over the area of interest for the radiometric bands green and NIR, the normalized difference, the Ostu threshold and finally creates a STAC catalog and items for the generated results.
This scenario is depicted below:
- read staged Landsat-9 data as a STAC Catalog and a STAC item, applies the crop over the area of interest for the radiometric bands green and NIR, the normalized difference, the Ostu threshold and finaly creates a STAC catalog and items for the generated results.
This scenario is depicted below:
Alice packages the application as an Application Package to include a macro workflow that reads the list of Sentinel-2 STAC items references or Landsat-9 staged data, launches a sub-workflow to detect the water bodies and creates the STAC catalog:
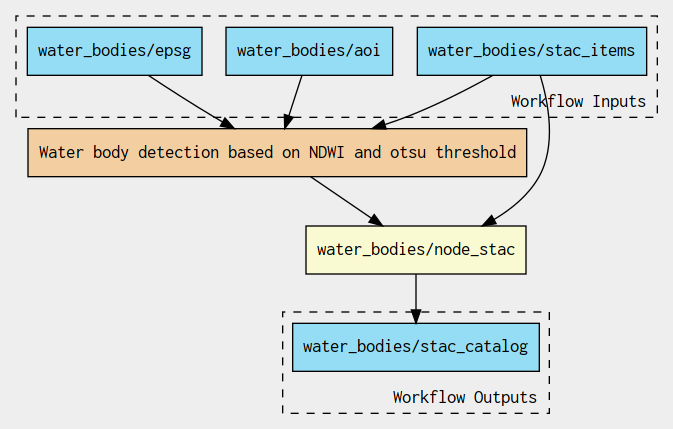
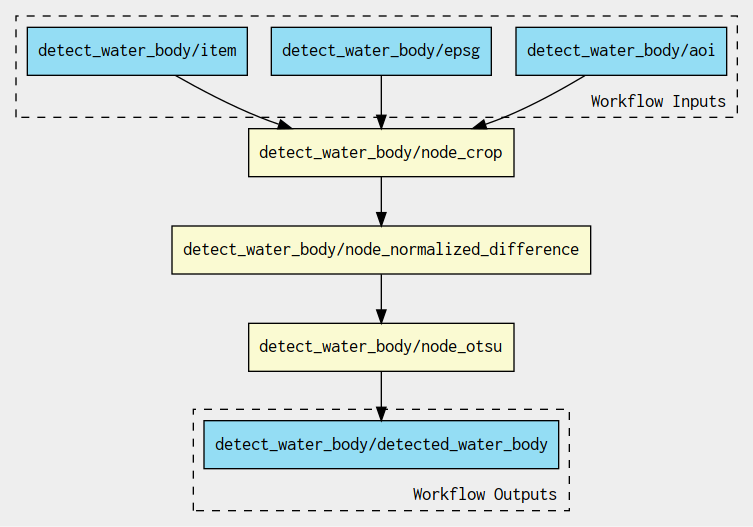
The sub-workflow applies the crop, Normalized difference, Otsu threshold steps:
The development and test dataset is made of two Sentinel-2 acquisitions:
| Acquisitions | ||
|---|---|---|
| Mission | Sentinel-2 | Sentinel-2 |
| Date | 2022-05-24 | 2021-07-13 |
| URL | S2B_10TFK_20210713_0_L2A | S2A_10TFK_20220524_0_L2A |
| Quicklook |  |
 |
And one Landsat-9 acquisition:
| Acquisition | |
|---|---|
| Date | 2023-10-15 |
| URL | LC09_L2SP_042033_20231015_02_T1 |
| Quicklook |  |
Each Command Line Tool step such as crop, Normalized difference, Otsu threshold and Create STAC runs a simple Python script in a dedicated container.